Dynamic Systems Analysis
To quantitatively understand and characterize cellular networks, a dynamic analysis of processes such as cell regulation is critical. However, our knowledge about essentially all cellular networks is incomplete, both regarding quantitative parameters and functional relations between components. We aim to employ dynamic mathematical models to infer such unknown quantities, for example, to allow for predictions across scales from the molecular level to patient responses in biomedical applications. The inference closely iterates between computation and experimentation, and we develop methods in both domains as needed.
Network Inference
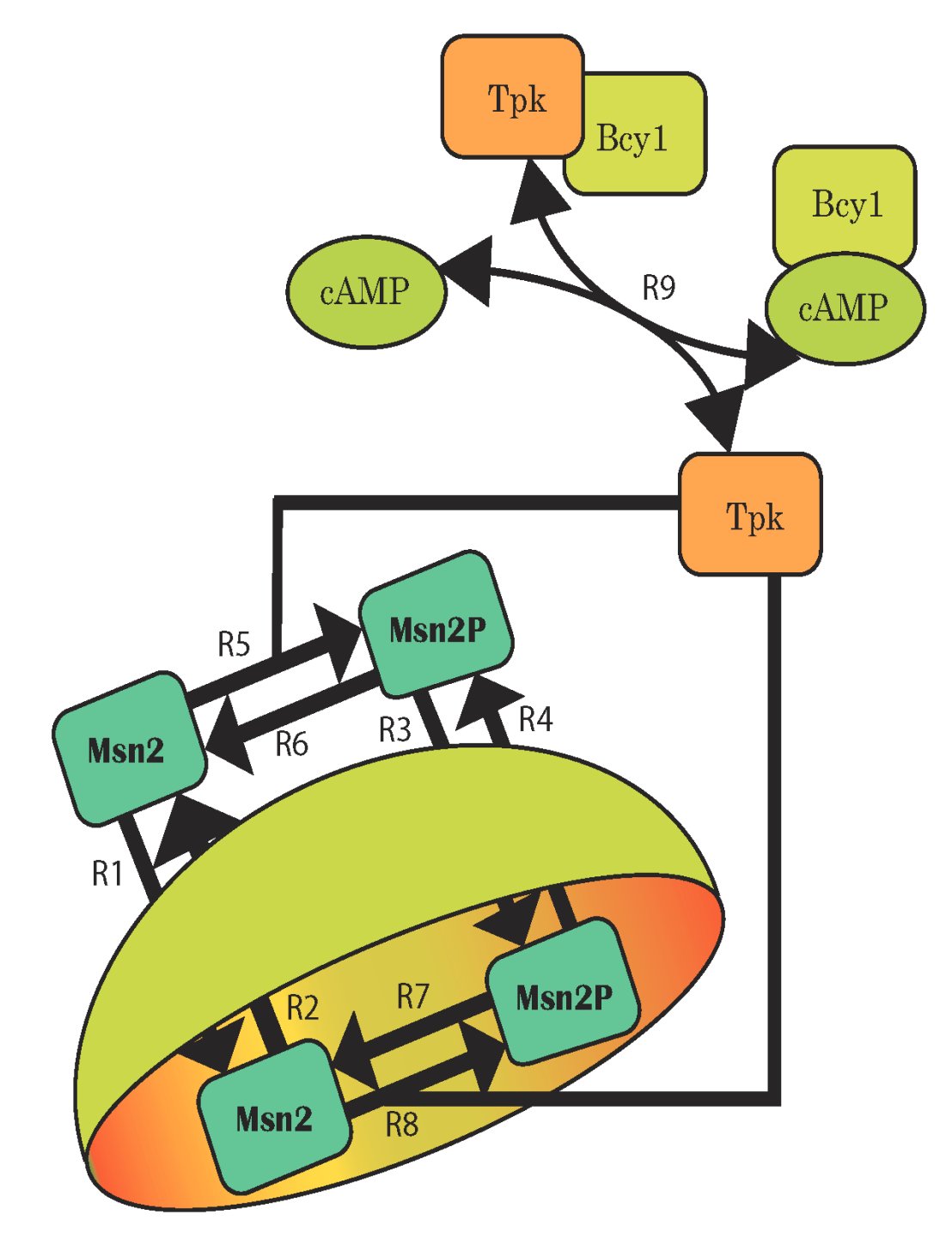
Uncertainty in components and their interactions – which leads to uncertain model structures – is a particular challenge for systems biology. This applies especially when mechanistic descriptions such as ordinary differential equation (ODE) models are needed. In the SystemsX.ch projects external pageYeastXcall_made and external pageSignalXcall_made, among others, we devise and apply scalable computational methods for the inference of mechanistic dynamic models. Our method of ‘topological filtering’ operates on ensembles of models with different structure and performs Bayesian inference via successive model reduction; its application to nutrient signaling in yeast revealed detailed control mechanisms. Other methods rely on stochastic noise to represent model uncertainties, or on coarse-grained identification approaches similar to frequency-domain analysis in control engineering.
Sunnaker M, Zamora-Sillero E, Lopez Garcia de Lomana A, Rudroff F, Sauer U, et al. (2014) Topological augmentation to infer hidden processes in biological systems. Bioinformatics 30: 221-227. external pagehttp://doi.org/10.1093/bioinformatics/btt638call_made.
Lang M, Stelling J (2014) Structural identification of nonlinear dynamic biomolecular feedback and feedforward loops. IFAC World Congress 2014. pp. 796-802. external pagehttp://doi.org/10.3182/20140824-6-ZA-1003.00357call_made.
Sunnaker M, Zamora-Sillero E, Dechant R, Ludwig C, Busetto AG, et al. (2013) Automatic generation of predictive dynamic models reveals nuclear phosphorylation as the key Msn2 control mechanism. Sci Signal 6: ra41. external pagehttp://doi.org/10.1126/scisignal.2003621call_made.
Kuepfer L, Peter M, Sauer U, Stelling J (2007) Ensemble modeling for analysis of cell signaling dynamics. Nat Biotechnol 25: 1001-1006. external pagehttp://doi.org/10.1038/nbt1330call_madeexternal page.call_made
Multiscale Modeling
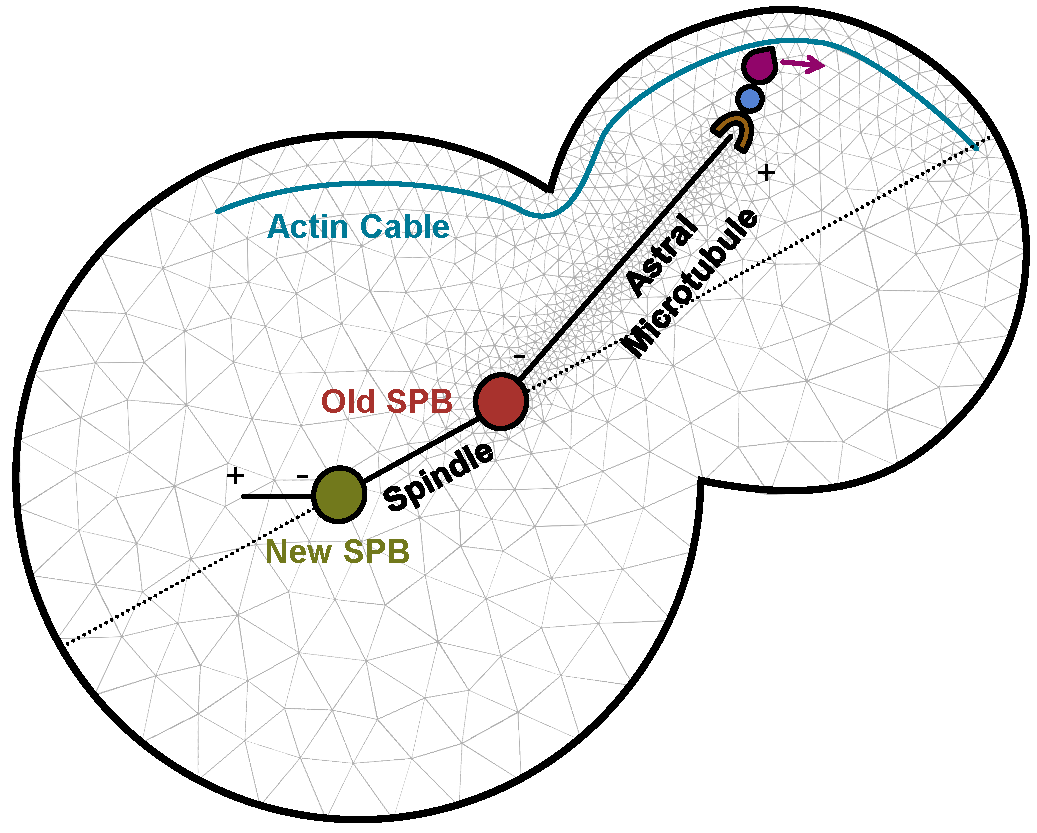
To address questions such as how a patient responds to a given drug, different time- and length-scales need to be covered. The challenges in model development are to define relevant scales, relevant and efficiently computable models at each scale, and suitable interfaces between the scales. Multiscale modeling is a central topic in several collaborative projects on, for example. spatial signaling and control of cellular architecture in yeast (external pageTubeXcall_made), influenza vaccination and drug development, and evasive resistance in cancer (external pageMERICcall_made).
Egli A, Humar A, Widmer LA, Lisboa LF, Santer DM, et al. (2015) Effect of Immunosuppression on Th2 and B-cell Responses to Influenza Vaccination. J Infect Dis. external pagehttp://doi.org/10.1093/infdis/jiv015call_made.
Stelling J, Kholodenko BN (2009) Signaling cascades as cellular devices for spatial computations. J Math Biol 58: 35-55. http://doi.org/10.1007/s00285-008-0162-6.
Robustness
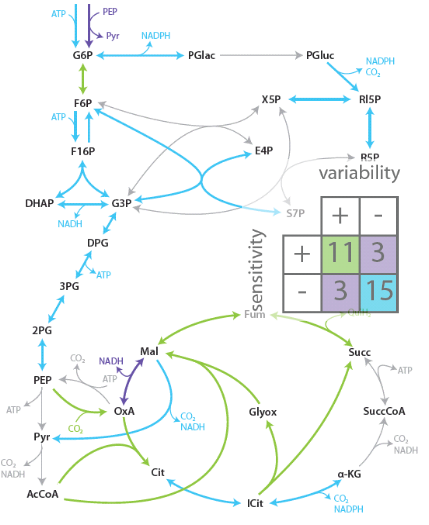
Robustness is the ability of a system to maintain performance in the face of perturbations and uncertainty. It is a long-recognized key property of living systems and also important for realizing reliable novel functions in synthetic biology. Because of its intimate link to network complexity, however, it is often not easy to identify causes of robustness. We employ model-based analysis methods to analyze this interplay between complexity and robustness. For example, our long-standing collaboration with the external pageDoyle group, UC Santa Barbaracall_made, aims at understanding mechanisms of robust performance in the circadian clock, the generator of daily rhythms in many organisms.
Mirsky HP, Taylor SR, Harvey RA, Stelling J, Doyle FJ (2011) Distribution-based sensitivity metric for highly variable biochemical systems. IET Syst Biol 5: 50. external pagehttp://doi.org/10.1049/iet-syb.2009.0064call_made.
Muller D, Stelling J (2009) Precise regulation of gene expression dynamics favors complex promoter architectures. PLoS Comput Biol 5: e1000279. external pagehttp://doi.org/10.1371/journal.pcbi.1000279call_made.
Bagheri N, Stelling J, Doyle FJ, 3rd (2007) Quantitative performance metrics for robustness in circadian rhythms. Bioinformatics 23: 358-364. external pagehttp://doi.org/10.1093/bioinformatics/btl627call_made.
Stelling J, Sauer U, Szallasi Z, Doyle FJ, 3rd, Doyle J (2004) Robustness of cellular functions. Cell 118: 675-685. external pagehttp://doi.org/10.1016/j.cell.2004.09.008call_made.
Numerical Methods
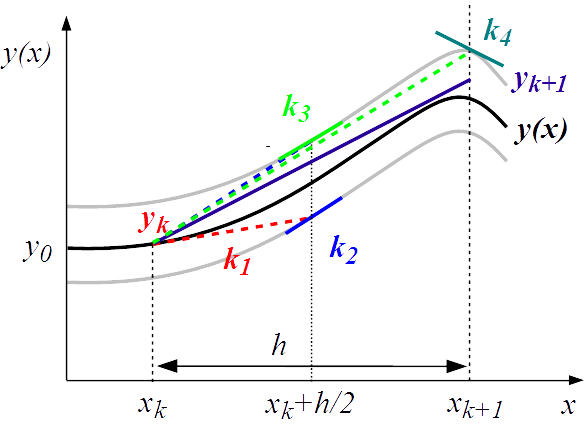
To address problems such as model-based inference at larger scale, current numerical computation methods are often not sufficiently efficient. We therefore developed several methods with more general applicability: odeSD, for the simultaneous integration of ODEs and parameter sensitivities; HYPERSPACE, co-developed with the external pageWagner group at Univ. Zurichcall_made, for sampling high-dimensional parameter spaces for dynamic systems; and an SDE evaluation package for evaluating parameter points in stochastic differential equation models.
Gonnet P, Dimopoulos S, Widmer L, Stelling J (2012) A specialized ODE integrator for the efficient computation of parameter sensitivities. BMC Syst Biol 6: 46. external pagehttp://doi.org/10.1186/1752-0509-6-46call_made.
Zamora-Sillero E, Hafner M, Ibig A, Stelling J, Wagner A (2011) Efficient characterization of high-dimensional parameter spaces for systems biology. BMC Syst Biol 5: 142. external pagehttp://doi.org/10.1186/1752-0509-5-142call_made.
Zak DE, Stelling J, Doyle FJ (2005) Sensitivity analysis of oscillatory (bio)chemical systems. Computers & Chemical Engineering 29: 663-673. external pagehttp://doi.org/10.1016/j.compchemeng.2004.08.021call_made.
Experimental Design
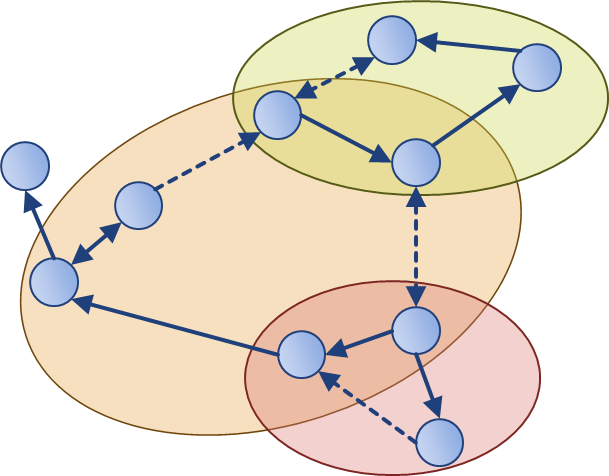
To enable network inference, it is essential that corresponding experiments are informative for the biological hypotheses to be discriminated. Optimal experimental design traditionally improves parameter estimation by selecting measurement variables, time points, and inputs locally with respect to existing models. Together with the Buhmann group (D-INFK), we proposed methods for near-optimal experimental design that rely less on model accuracy. Our approach for modular experimental design requires only the network structure, and thereby connects structural network analysis and dynamic systems.
Lang M, Summers S, Stelling J (2014) Cutting the wires: modularization of cellular networks for experimental design. Biophys J 106: 321-331. external pagehttp://doi.org/10.1016/j.bpj.2013.11.2960call_made.
Busetto AG, Hauser A, Krummenacher G, Sunnaker M, Dimopoulos S, et al. (2013) Near-optimal experimental design for model selection in systems biology. Bioinformatics 29: 2625-2632. external pagehttp://doi.org/10.1093/bioinformatics/btt436call_made.
Imaging and Image Analysis
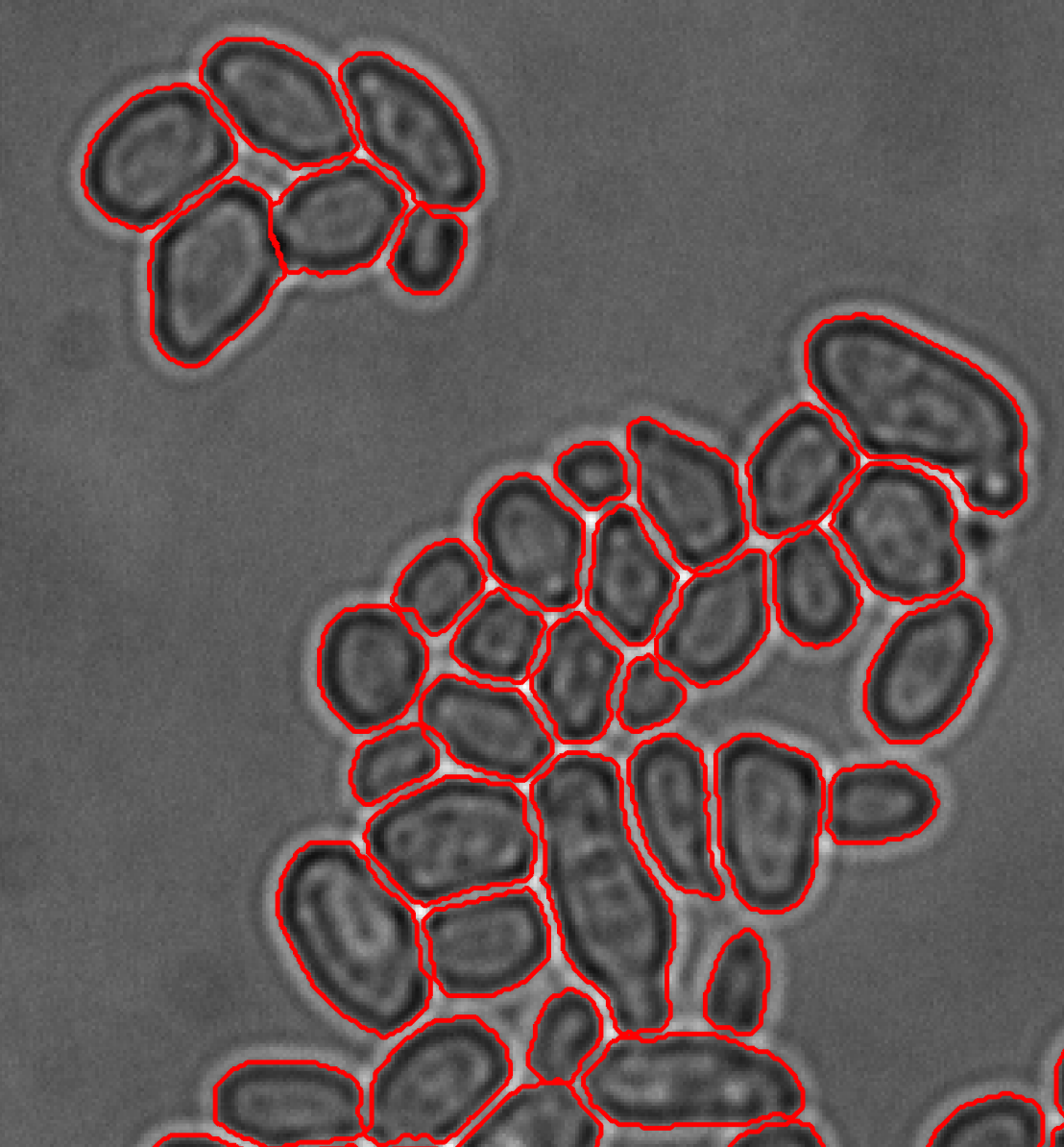
Our activities in yeast experimental biology focus on quantitative imaging approaches for data generation. To better link model-based and experimental systems analysis, we developed software tools for advanced microscopy. The high level, open-source microscope control platform external pageYouScopecall_made allows users to implement complex measurement protocols on motorized microscopes to match the capabilities of experimental design approaches. CellX is an open-source software package for cell segmentation, intensity quantification, and cell tracking to quantify features of individual cells using light microscopy for diverse biological samples and imaging techniques.
Dimopoulos S, Mayer CE, Rudolf F, Stelling J (2014) Accurate cell segmentation in microscopy images using membrane patterns. Bioinformatics 30: 2644-2651. external pagehttp://doi.org/10.1093/bioinformatics/btu302call_made.
Mayer C, Dimopoulos S, Rudolf F, Stelling J (2013) Using CellX to quantify intracellular events. Curr Protoc Mol Biol Chapter 14: Unit 14 22. external pagehttp://doi.org/10.1002/0471142727.mb1422s101call_made.
Lang M, Rudolf F, Stelling J (2012) Use of YouScope to implement systematic microscopy protocols. Curr Protoc Mol Biol. pp. Unit 14 21 11-23. external pagehttp://doi.org/10.1002/0471142727.mb1421s98call_made.
