Research
The main research theme of the CSB group is the development of theoretical, computational, and experimental methods for dealing with uncertainty in the analysis and design of complex, potentially large-scale biological networks, and their application to address fundamental problems in systems and synthetic biology. We combine computational and theoretical approaches based on mechanistic mathematical models with biological experimentation, either by studying yeast as a model organism within the group, or in various external collaborations. The research aims and methods define our four overlapping research areas.
Structural Network Analysis
Structures (topologies) of biological networks are often much better characterized than detailed interaction mechanisms. In structural network analysis, we aim at elucidating functional features from topologies alone, for example, to predict the behavior of large-scale metabolic networks or to infer cellular regulation. Read more
Dynamic Systems Analysis
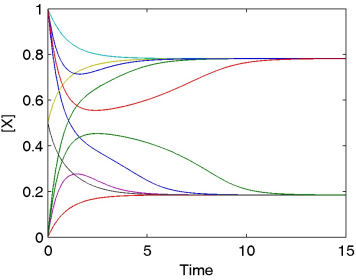
Detailed systems dynamics are, for example, required to understand the integration of cellular regulation across metabolism, signal transduction, and gene regulation. We aim at developing advanced methods and models for detailed network analysis that cope with the prevailing uncertainty due to a lack of experimental data and biological knowledge. Read more
Synthetic Biology
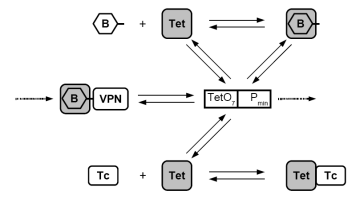
The rational design of biological circuits with novel functions is a key challenge of synthetic biology. We aim at establishing model-based design methodologies to eventually allow for automatic design and implementation of large-scale circuits, especially in view of medical applications of artificial gene circuits that perform reliably. Read more
Experimental Yeast Biology
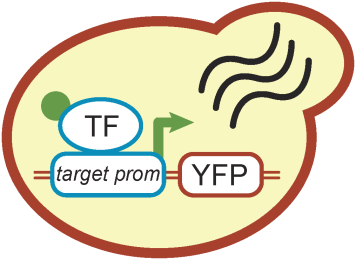
The experimental part of the group complements the theoretical efforts by using budding yeast as a model organism. Often relying on imaging based analysis and classical biochemistry, we particularly aim at the quantitative study of cell signaling and growth control, and the efficient design and implementation of synthetic gene circuits in eukaryotes. Read more
